Automatic EEG sleep stage scoring
One third of every person's life is spent sleeping (Liang et al., 2012). Sleep deprivation can significantly impact human health, wellbeing and quality of life and may result in co-morbidities and even mortality (Hublin et al., 2007). Human newborns spend two-thirds of their lives sleeping. The quality of sleep plays an important role in infant neurodevelopment especially in the first year of life. In infants, it is very difficult to study brain functions through cognitive examinations. This is why sleep studies can be very useful to investigate the neurodevelopmental process and brain abnormalities during the infancy period by exploring sleep structure. In sleep studies, electroencephalography (EEG) is routinely used as an efficient tool to systematically investigate sleep structure and stages. Each sleep stage is identified with a set of specific physiological markers accompanied by characteristics changes in spectral features, which are used to explore the sleep architecture including the timing and organization of sleep stages over the course of sleep.
According to R&K guidelines (Rechtschaffen and Kales, 1968), a healthy adult periodically passes through six sleep stages including wakefulness (W), rapid eye movement (REM) and non-rapid eye movement stages (NREM, further classified into stages S1-S4), at intervals of around 90-120 min over a whole night (Carskadon and Dement, 2005; Chokroverty, 2017). Normal sleep cycles typically begin with NREM light sleep stages (S1 and S2) and then progress into deeper stages (S3 and S4) and REM sleep (Doroshenkov et al., 2007). The American Academy of Sleep Medicine (AASM) scoring manual recognizes five sleep stages, wakefulness (W), stage N1 (S1 in R&K), stage N2 (S2 in R&K), stage N3 (S3 and S4 in R&K), and stage REM.
Compared to adults, neonates show markedly inhomogeneity and instability in EEG during wake or sleep. In neonates, two major sleep states are clearly distinguished, the Quiet Sleep (QS equivalent to non-REM sleep in adults) and Active Sleep (AS equivalent to REM sleep), with in-between indeterminate or transitional epochs exhibiting non-EEG characteristics of AS and EEG characteristics of QS. The transitional epochs are often observed at sleep onset (arousal state) and between REM and non-REM sleep. Overall, term neonates spend 50-60% of their sleep cycles in AS, 30-40% in QS, and 10-15% in the transitional state, with sleep-wake and sleep-only cycles lasting typically 3-4 hours and 40-70 min, respectively.
Visual sleep stage scoring is performed by trained neurologists, a time-consuming and costly procedure that is highly prone to human errors depending on experts’ experiences. Automatic sleep staging can achieve high accuracy and efficiently reduce scoring time. In two studies, we developed single-channle and multichannel methods for EEG sleep stage scoring in adults and full-term neonates, respectively.
Single-channel EEG sleep stage scoring in adults
In the first study, we developed an efficient single-channel sleep stage scoring tool with improved accuracy in comparison with other existing methods. In the first stage of the tool, an efficient feature selection method called MGCACO (Modified Graph Clustering Ant Colony Optimization) (Ghimatgar et al. 2018) was used to select optimal feature sets from a pool of characteristic features based on the relevance and redundancy analysis. In the second stage, a random forest classifier, shown to be highly resistant to overfitting and missing data problems, was used for sleep stage classification (Hassan and Bhuiyan, 2016a; Boostani et al., 2017; JIANG et al. 2019). In the last stage, an efficient postprocessing method was developed based on HMM to incorporate the temporal structure of sleep cycles into the sleep stage scoring procedure to reduce false positives resulted from the classification stage (Fig. 1).
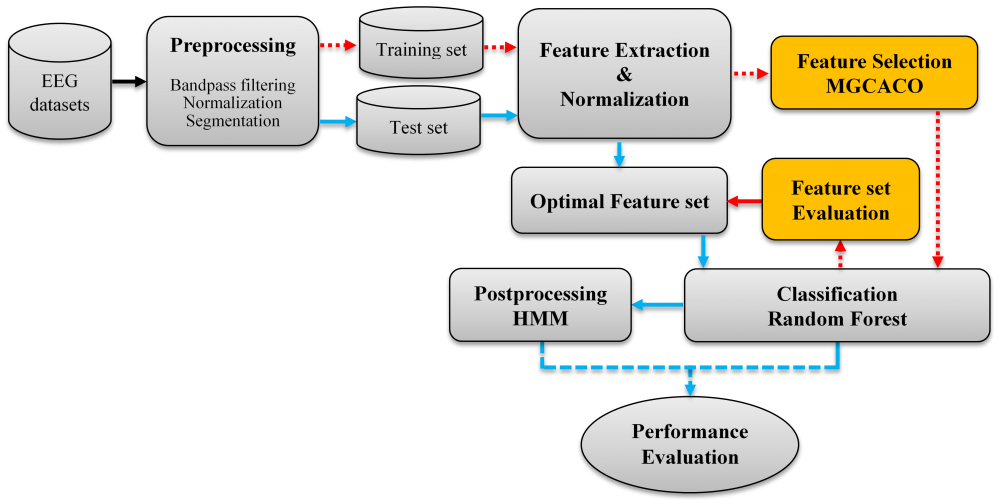
Figure 1. Block diagram of the proposed algorithm for EEG-based sleep stage scoring in adults.
We evaluated the proposed algorithm on four publicly available sleep EEG datasets, EDF sleep database, EDF sleep database (Expanded), Dreams Subject Database and ISRUC (subgroup 3) database (Ronzhina et al., 2012; Imtiaz and Rodriguez-Villegas, 2014; Hassan and Bhuiyan, 2016a; Sharma et al., 2017; Khalighi et al., 2016). After preprocessing, from each 30-sec EEG segment, a set of 136 features was extracted mostly in the time and frequency domain (Aarabi et al., 2006; Aarabi et al., 2007). The features were roughly grouped into six categories including autoregressive coefficients and time, frequency, wavelet, cepstral and nonlinear features. We then used MGCACO, a feature selection method based on the relevance and redundancy analysis to substantially reduce the computational complexity and improve the classifier perfomance. We then used the random forest, a classifier based on the combination of multiple decision trees and the majority voting. We finally used two first-order fully-connected HMMs to incorporate the sleep temporal structure into the sleep stage scoring procedure based on the R&K and AASM rules (Fig. 2). In this model, transitions between states are described probabilistically. We adopted the HMM for sleep stage scoring based on the hypothesis that the occurrence of a given stage can be influenced by the occurrence of its previous stage.
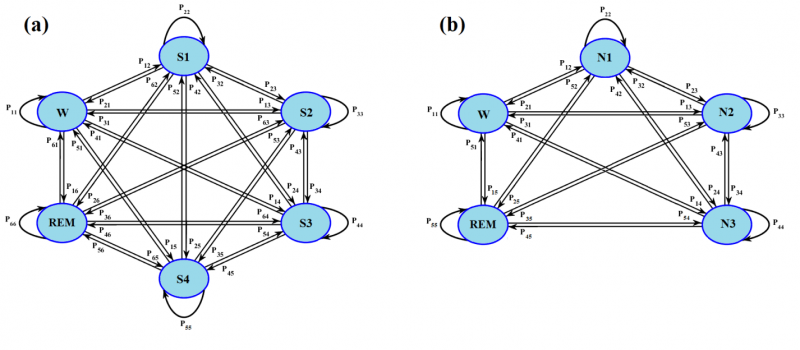
Figure 2. Fully connected HMM models used for refining the classification results obtained based on the R&K (a) and AASM (b) scoring rules.
Using the leave-one-out validation strategy, our method achieved overall accuracies in the range of (79.4-87.4%) and (77.6-80.4%) with Kappa values in the range of 0.7–0.85 for the six-stage (R&K) and five-stage (AASM) classification cases, respectively. Our method showed a reduction in overall accuracy up to 8% using the cross-dataset validation strategy in comparison with the subject cross-validation method. Fig. 3 shows the average HMM optimized based on the R&K and AASM guidelines, respectively. As shown, the transition probabilities between (S3 and S4) are relatively strong, suggesting that stages S4 and S3 should be truly combined as defined in the AASM scoring rules. These models further illustrate infrequent sleep stage transitions that are less likely to occur in normal healthy subjects. We used the average HMM for each dataset to reduce false positives of the sleep stage classification.
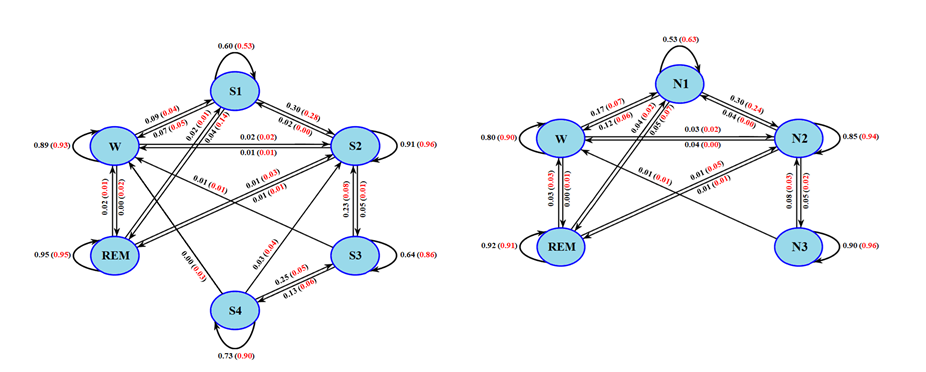
Figure 3. Average HMMs optimized based on the automatic scoring performed by the proposed method (with probabilities shown in black) in comparison with the expert scoring (with probabilities shown in red).
Multi-channel EEG sleep stage classification in full-term neonates
In neonates, automatized EEG sleep staging is more challenging due to the nonstationary and nonlinear nature of EEG signals as well as asymmetry between brain regions. This is why single-channel methods provide relatively lower accuracy rates compared to multi-channel methods developed for sleep staging in neonates. In the second study, we developed an automated multichannel classification method for EEG sleep staging in full-term neonates (Fig. 4). To improve the prediction performance and reduce the computational complexity, the method relied on a four-stage processing pipeline – feature extraction and selection, channel selection, classification, and postprocessing. In the first stage after preprocessing, a comprehensive set of time, frequency, and nonlinear features widely used in sleep studies in adults and neonates was extracted from EEG segments. The MGCACO method was then used to select optimal feature sets based on the relevance and redundancy analysis. A Bidirectional Long Short-Term Memory (BiLSTM) network was trained using the optimal feature sets and used for sleep stage classification. To improve the classification performance, a channel selection method was used to optimize the spatial sequence of EEG channels. Finally, in the postprocessing stage, an HMM was employed to incorporate the temporal organization of sleep cycles into the sleep stage classification procedure. The performance of the method was evaluated using the the K-fold (KFCV) and LOOCV strategies on sleep EEG data from full-term healthy neonates .
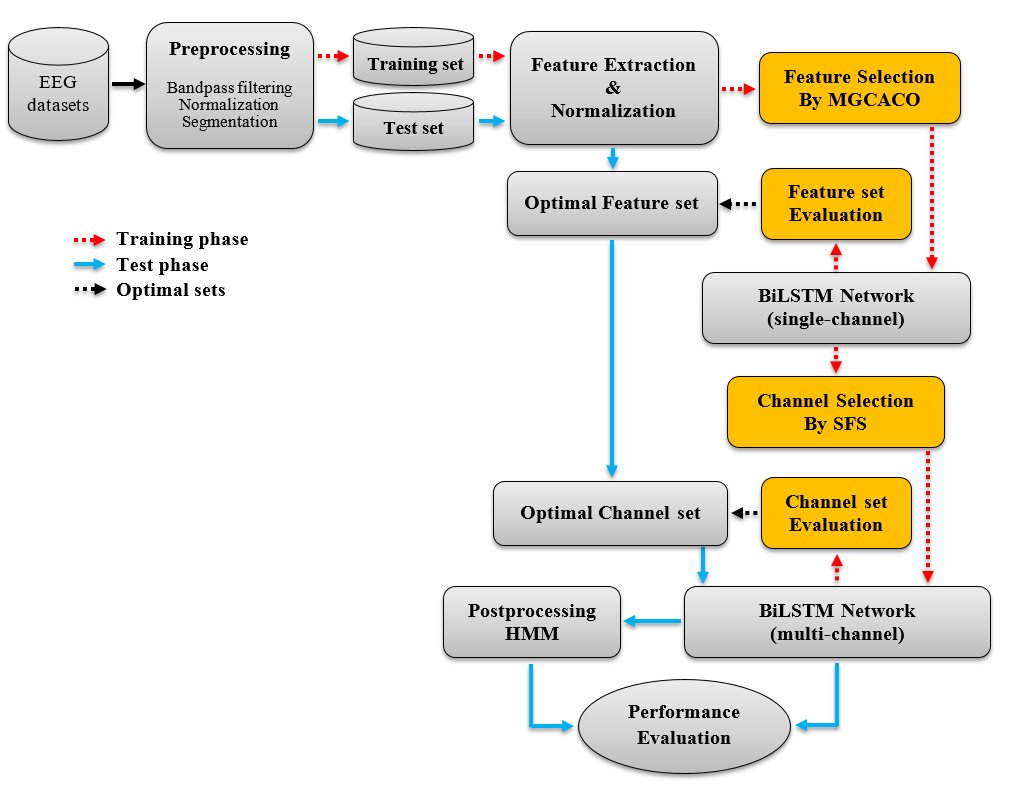
Figure 4. Block diagram of the automated algorithm designed for EEG sleep stage scoring in neonates.
We used the multichannel EEG sleep data recorded from 16 full-term healthy neonates of 38–40 weeks postmenstrual age at the neonatal intensive care unit (NICU) of the University Hospitals, Leuven, Belgium for method development and evaluation. The manual staging has been visually performed by two EEG experts, who identified four sleep stages Active Sleep 1 (ASI), Active Sleep 2 (ASII, LVI), Quiet Sleep 1 (HVS) and Quiet sleep 2 (TA) as well as two other states defined as indeterminate (IS) and artifacts.
After preprocessing, a comprehensive set of 168 features widely used in sleep studies in adults and neonates was extracted from each 30-sec EEG segment. To reduce the risk of overfitting caused by redundancy between features, we used MGCACO, an efficient multivariate filter-based feature selection approach, for dimensionality reduction. We further used a channel selection method to improve the classification performance and reduce the computational complexity by identifying brain areas more relevant to sleep staging in neonates. We then used the BiLSTM network, a powerful Recurrent Neural Network (RNN) to capture long-range dependencies between multichannel EEG signals. Finally, We used an HMM to model the sequential nature of the sleep stages (ASI, LVI, HVS and TA) labeled by the classifier (Fig. 5).
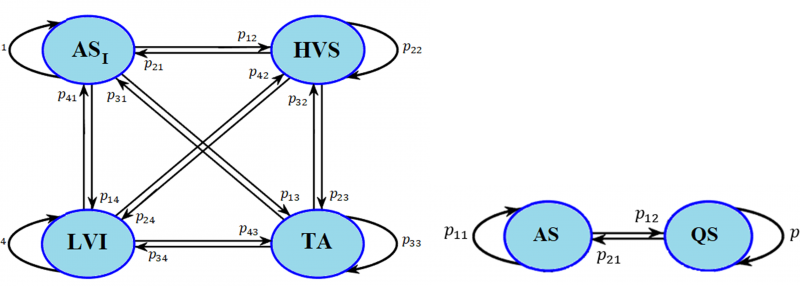
Figure 5. Fully connected HMM used for postprocessing for the four-class and two-class cases. ASI: active sleep (sub-state 1), LVI: Low-Voltage Irregular, TA: Tracé Alternant, HVS: High Voltage Slow-wave.
Our method achieved a mean kappa and an overall accuracy of 0.71 (0.76) and 78.9% (82.4%) using the bipolar montage and the KFCV (LOOCV) strategy. In comparison with the bipolar montage, the monopolar montage led to lower accuracies. In comparison with the multi-channel approaches evaluated using the LOOCV strategy, our results achieved significant improvements in accuracy up to 9% and 12% for the two-class and four-class cases, respectively.
Compared to forty features selected in our previous study for sleep stage classification in adults, sleep scoring in neonates required twenty more features (up to sixty features in total) to improve the performance and reach stability in accuracy. In our algorithm, the channel selection procedure also reduced the computational complexity and improved the classification performance by 2%. Our results confirm the superiority of the multichannel approaches over the single-channel methods for EEG-based sleep staging in neonates.
Finally, the average optimized HMMs (Fig. 6) obtained for the four-state and two-state cases showed stronger transition probabilities between (HVS and TA) and (ASI and HVS). In the four-state model, the infrequent sleep stage transitions with probabilities weaker than 0.01 (1%) were eliminated during the optimization process. Overall, the HMM-based postprocessing stage could improve the overall accuracy by 3% for all stages by reducing false positives observed mostly for HVS (monopolar montage) and LVI (bipolar montage).
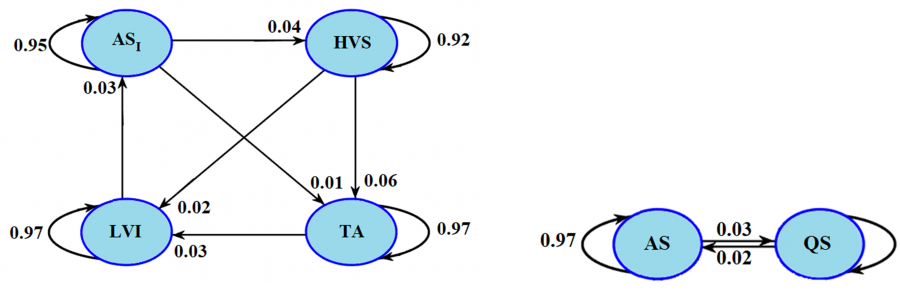
Figure 6. Hidden Markov models optimized for the four-state and two-state cases using the LOOCV strategy. In the four-state model, inter-state transitions with probabilities weaker than 0.01 (1%) were considered as infrequent transitions and eliminated. ASI: active sleep (sub-state 1), LVI: Low-Voltage Irregular, TA: Tracé Alternant, HVS: High Voltage Slow-wave.
